
© istock.com/kickers
P roteins residing in fossils are rarely ready to analyze immediately after paleontological digs. Instead, researchers have to put samples through a series of steps designed to break down surrounding material and to solubilize and separate out the proteins. Only then can the peptides be sequenced, a process generally carried out using mass spectrometry. To minimize the risk of contamination of samples with external peptides—a particular concern for ancient samples given that endogenous proteins are usually present in low abundance—researchers prepare controls at every step of the analysis.
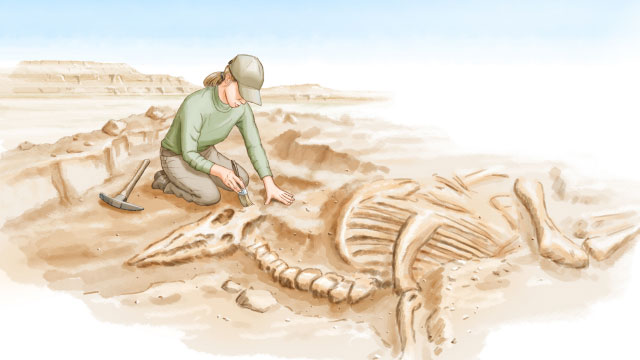
1. Collection
 © GEORGE RETSECK
© GEORGE RETSECK
The starting point for many projects in paleoproteomics is a paleontological site. In the right conditions, proteins can survive for hundreds of thousands, potentially millions, of years, before completely succumbing to the ravages of degradation. Fossilized bones that have been preserved in dry ground, for example, may harbor structural proteins such as collagen and...
Contamination control: Researchers take samples of the soil or sediments that surround fossils in order to eliminate any proteins that are not endogenous to the ancient material being studied from subsequent analyses.
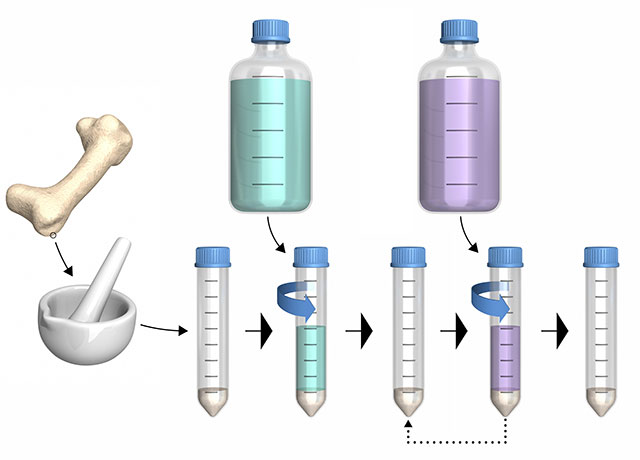
2. Preparation
 © GEORGE RETSECK
© GEORGE RETSECK
The fossil is ground into a powder and then treated with various chemicals to separate the proteins from the surrounding material. First, researchers add demineralizing agents such as hydrochloric acid or ethylenediaminetetraacetic acid (EDTA) to free proteins from minerals in fossils. Second, researchers add solubilizing agents such as ammonium bicarbonate or guanidine hydrochloride. The resulting solution is then put through multiple rounds of centrifugation and resolubilization before the proteins are analyzed by mass spectrometry.
Contamination control: Researchers simultaneously perform the above steps with a buffer solution. Unexpected proteins discovered in these controls are likely to be from human investigators—keratin from hair or clothes, for example—and can be flagged in later analyses.
3. Analysis
 © GEORGE RETSECK
© GEORGE RETSECK
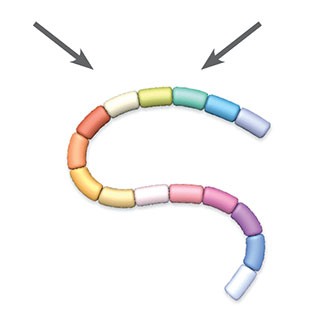 © GEORGE RETSECKMost research groups analyze ancient proteins using mass spectrometry—an approach that determines the molecular makeup of a compound by analyzing the mass and charge of its particles. For identification, proteins are typically digested into smaller peptides by chemicals or enzymes. Inside a tandem mass spectrometer, one of several types of mass spectrometer, these peptides are then ionized to give them a positive charge and accelerated through a specialized chamber, with lighter molecules accelerating more quickly than heavier ones. The peptides are measured by a mass analyzer before being broken into even smaller fragments and measured by a second mass analyzer. These measurements produce two sets of mass spectra that can then be compared against a database for protein identification or used to help reveal the sequence of amino acids making up the original peptide.
© GEORGE RETSECKMost research groups analyze ancient proteins using mass spectrometry—an approach that determines the molecular makeup of a compound by analyzing the mass and charge of its particles. For identification, proteins are typically digested into smaller peptides by chemicals or enzymes. Inside a tandem mass spectrometer, one of several types of mass spectrometer, these peptides are then ionized to give them a positive charge and accelerated through a specialized chamber, with lighter molecules accelerating more quickly than heavier ones. The peptides are measured by a mass analyzer before being broken into even smaller fragments and measured by a second mass analyzer. These measurements produce two sets of mass spectra that can then be compared against a database for protein identification or used to help reveal the sequence of amino acids making up the original peptide.
Contamination control: Researchers analyze control samples to test for the possibility that some protein fragments have been left inside the mass spectrometer from a previous experiment. Some groups also use separate machines for old and new samples to reduce the possibility of accidental detection of modern proteins in ancient material.
Read the full story.
Interested in reading more?





