ABOVE: © ISTOCK.COM, KTSIMAGE
About 700 million years ago, sponges branched off from all other animals on the tree of life. Despite this evolutionary distance, sponges share a form of gene regulation with much more complex species. The mechanisms are so similar, in fact, that a genetic element called an enhancer from the sea sponge Amphimedon queenslandica can drive transcription in specific cell types in mice and zebrafish, despite the fact that the genomes of these animals don’t normally include a similar sequence, according to a study published late last year in Science.
The result was totally surprising, says Emily Wong, a computational genomics researcher at Victor Chang Cardiac Institute in Australia and a coauthor of the study. “We didn’t think it would work.”
The results serve as an extreme example of what scientists are now recognizing as trends among enhancers—that activity can be conserved over long evolutionary timescales and that such conserved activity doesn’t require DNA sequences to match perfectly. “Just the sheer distance between the species . . . makes that [gene regulatory activity] really exciting and cool,” says Tony Capra, an evolutionary geneticist at the University of California, San Francisco, who was not involved in the research.
It is really complicated. And that, of course, makes it interesting and fun.
—Paul Flicek, European Molecular Biology Laboratory
While there are some examples of “ultraconserved” enhancer elements that are identical in rodents and humans, increased genome sequencing has revealed that enhancers often evolve rapidly, accumulating substantial sequence changes over relatively short periods. Yet enhancers have maintained molecular relationships with the proteins that regulate transcription, many of which have been preserved over hundreds of millions of years. The rules underlying how enhancer sequences interact with these proteins to modulate transcription remain murky, and scientists are still sorting out the details of enhancer biology and trying to understand how function is conserved even as the DNA code mutates.
“It is really complicated,” says Paul Flicek, a computational biologist at the European Molecular Biology Laboratory’s European Bioinformatics Institute in the UK. “And that, of course, makes it interesting and fun.”
Enhancer grammar
Enhancers contain transcription factor (TF) binding sites and regulate transcription at a distance—sometimes a very great distance. In mice and humans, for example, an enhancer element for the Shh gene, involved in patterning during embryonic development, is located about 1 million base pairs away, and mutations in this enhancer element can cause organisms to develop extra fingers or toes.
Beyond those universal qualities, enhancers vary dramatically. They can be found upstream or downstream of genes, or even within them. Enhancers also vary in length, ranging from about 10 to 1,000 base pairs, and contain different numbers of TF binding sites. TFs glom on to the DNA and recruit components of the transcriptional machinery to promoters, stretches of the genome where gene transcription is initiated. How exactly TFs assemble at enhancers to influence gene regulation remains a bit of a black box, however.
One idea, dubbed the enhanceosome model, posits that a specific number of TFs and other proteins must be present in a defined orientation to influence gene expression. These rules are also known as enhancer grammar. A classic example of this model is the interferon-β enhancer, where eight proteins, including three TFs, must be present on the DNA to direct the transcriptional machinery to the interferon-β gene’s promoter. In an alternative “billboard” model, TF binding to enhancer sequences doesn’t rely on grammar: the order, number, and spacing can vary and still influence gene expression, sometimes in different ways. “It’s really that these are two ends of a spectrum,” says Emma Farley, a molecular biologist at the University of California, San Diego. A third concept, called the TF-collective model, adds another layer of complexity by proposing that transcription factors bound to DNA recruit additional TFs via protein-protein interactions; these extra TFs could influence transcription without binding defined DNA sequences. (See illustration below.)
All these models for enhancer function, ranging from rigid enhancer grammar to flexible grammar to no grammar, could be correct, depending on biological context. “We need to understand why different types of enhancers fall at different places on that spectrum,” Farley says. “That might help us understand the different nuances of enhancer grammar better.”
Because there’s no means of spotting enhancers by sequence alone, scientists identify regions of regulatory activity based on indirect readouts. For example, active enhancers are typically associated with open chromatin—stretches of DNA not tightly wound around nucleosomes. Another approach examines DNA associated with certain versions of histones, the proteins that make up nucleosomes. Characteristic patterns of histone modifications decorate enhancer regions. For both strategies, biochemical identification and sequencing of targeted areas can yield putative enhancers.
The results generated by such approaches don’t always agree, notes Capra, but “I don’t see that as necessarily a problem. I think it just reflects that we are studying a very complex biochemical process that has many inputs and outputs [and] many different signatures that it leaves along the genome over time.” That said, he adds in an email, “the disagreement is a problem when researchers do not account for it.”
Enhancer FunctionEnhancers mediate gene expression by recruiting transcription factors (TFs) that subsequently recruit additional machinery to initiate transcription. Enhancers often act across great distances in the genome—up to about 1 million base pairs, as is the case for the developmental patterning gene Shh—and their location relative to the genes they regulate is variable: they can be upstream or downstream, and they can reside outside of coding areas entirely or within introns of other genes.  © BODY SCIENTIFIC INTERNATIONAL |
Conservation of enhancer activity
As demonstrated by the sponge enhancer, conserved activity doesn’t rely on identical DNA sequences. But there are cases where evolution has preserved the exact sequences of enhancer elements in distantly related species. A striking example of how enhancers can be conserved at the sequence level are “ultraconserved” elements, first described in 2004 by collaborators at the University of California, Santa Cruz, and the University of Queensland in Australia. Their study uncovered 481 segments longer than 200 base pairs that were perfect matches across human, rat, and mouse genomes; later studies on 245 of these elements located in noncoding stretches of the human genome found that about half had enhancer activity and could drive gene expression during mouse development.
The discovery of ultraconserved elements was astounding considering that about 80 million years separate humans and rodents from our last common ancestor. The researchers calculated that, based on even a slow mutation rate, the probability that one such genetic sequence would by chance exist in the approximately 3 billion bases of the human genome would be less than 1 in 1022. Consequently, scientists assumed that ultraconserved sequences must be essential. But they were wrong: mice remained viable and fertile even when some of these enhancers were removed.
“People were really kind of shocked by it,” says Diane Dickel, a genomicist at Lawrence Berkeley National Laboratory who did not participate in that work. She was involved in later studies that found that removing ultraconserved enhancers did have an effect—it was just subtle. More recently, she and her colleagues mutated 23 ultraconserved enhancers and found that, for the most part, they still had activity during development. The findings suggest that even these highly conserved enhancers can still tolerate sequence changes, which begs the question of why more variation hasn’t crept in over millions of years. “We still don’t have a completely clear understanding about why these sites are so perfectly conserved,” Dickel says.
But ultraconserved elements are an exception in enhancer evolution. In a landmark study in 2015 that tracked promoters and enhancers across 20 mammalian species using histone marks, the researchers found that promoters evolved slowly while enhancers evolved rapidly.
Despite a lack of conservation in overall sequence, enhancers from distantly related species do have something in common: TF binding site sequences. Transcription factors are deeply conserved across the tree of life. A comparison of TFs in fruit flies, mice, and humans revealed that the different versions of the proteins had similar binding properties and recognized the same DNA sequences. This conservation could be partly why the A. queenslandica enhancer worked in mice and zebrafish despite hundreds of millions of years of evolutionary distance. While the enhancer sequences didn’t align overall in that study, common short TF binding motifs appeared in the sponge, mouse, and zebrafish enhancers, albeit in different arrangements.
Just the sheer distance between the species . . . makes that [gene regulatory activity] really exciting and cool.
—Tony Capra, University of California, San Francisco
Sequence similarity or not, “the fact that these enhancers work actually means that animals over these vast distances share what one might call a ‘cell type,’” defined by a common set of TFs, says Alexander Stark, a genomicist at the Research Institute of Molecular Pathology in Austria who was not involved in the sponge study. This shared cell type was apparently maintained across evolutionary time in distantly related species, he continues, and TFs recognize conserved binding motifs, despite their rearrangements in the different animal genomes.
“The relative position and orientation of these short motifs with respect to each other often doesn’t [seem to] matter,” Stark says. There are also some cases where it does. Experiments using engineered enhancers from the sea squirt Ciona intestinalis, for example, found that changes in the spacing or arrangement of binding motifs could compensate for weak binding sites to drive gene expression.
Evolution is acting on what genes are turned on and when, Capra says. “Evolution doesn’t really care what particular sequences along the genome are driving that activity. It’s putting selection on the output of gene expression.”
Another reason enhancer function is often conserved despite sequence changes is redundancy. These genetic elements can have multiple copies of TF binding sites, so changes to one site may not negatively affect transcription—especially with the billboard model. There may also be more than one enhancer acting on a gene. These built-in backups may allow mutations to accumulate without obvious functional consequences. But these qualities do make enhancers difficult to study, says Flicek.
Analyses of enhancers typically involve knocking out individual TF binding sites. But imagine a scenario with 20 people pushing a car up a hill, Flicek says. Removing one person may reduce efficiency slightly, but the car could still make it up the hill. You might conclude that the missing person wasn’t essential. If you went through this process with all 20 people, you could conclude that none of them were essential. “But that’s not what’s really happened,” Flicek says. “What’s really happened is there’s redundancy that we don’t see, and we can’t understand.”
Enhancer EvolutionEnhancers are stretches of DNA that regulate where and when a gene is expressed. While the sequences of enhancers can vary among species, their function is highly conserved across hundreds of millions of years of evolution. For example, a recent study found that an enhancer from the sponge Amphimedon queenslandica can drive transcription in specific cell types in mice and zebrafish. While enhancers in the more complex organisms didn’t match the sequence of the sponge enhancer, the regions contained different arrangements of shared transcription factor binding motifs. The same was also true in the human region that most closely matched the sponge enhancer. 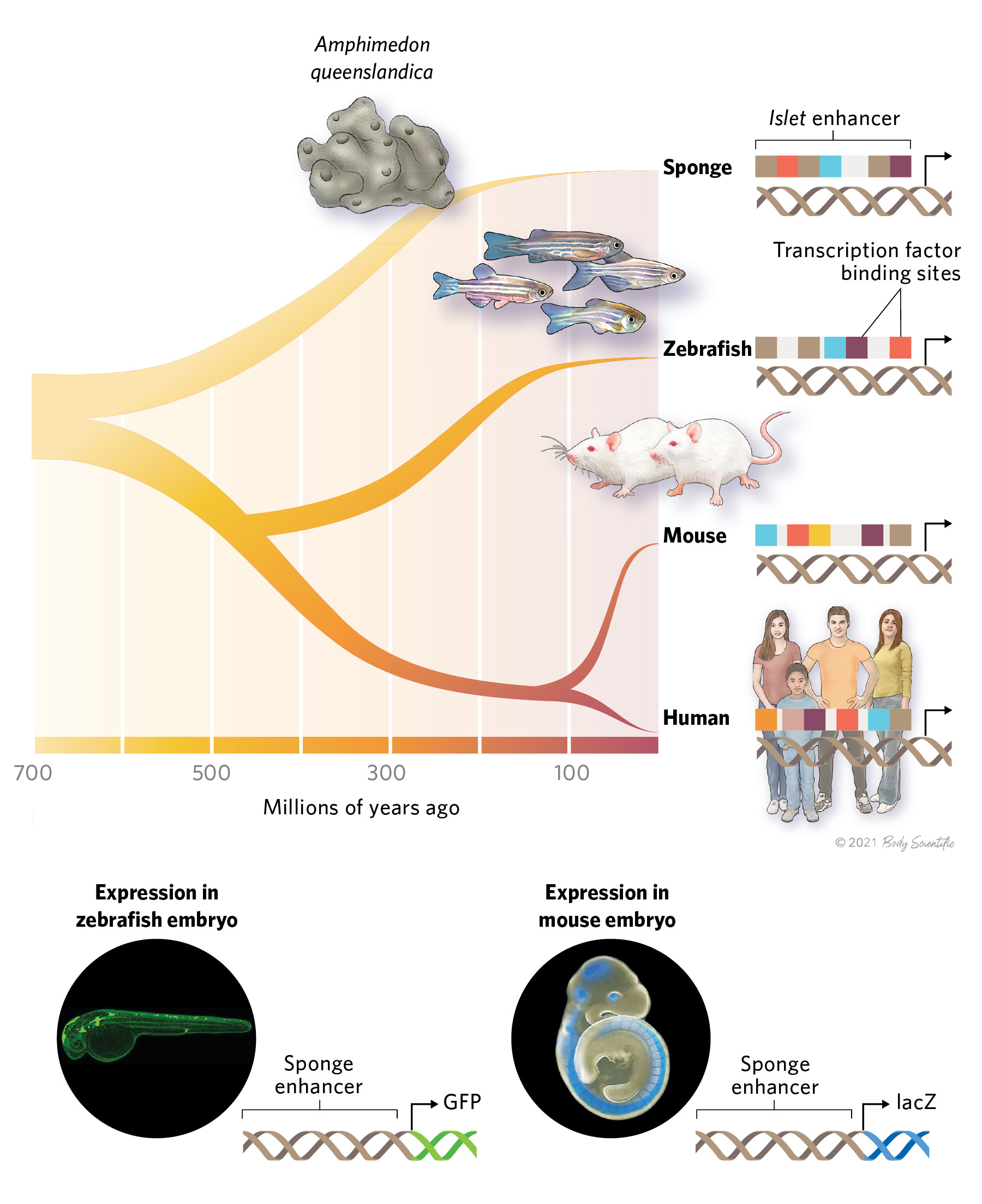 © BODY SCIENTIFIC INTERNATIONAL |
Enhancers over time
While most enhancers do evolve quickly, there are still constraints on how much they change. The A. queenslandica enhancer that drove activity in mice and zebrafish, for example, is situated within a bystander gene separate from the one whose expression it modified. Such an arrangement could limit how much the enhancer can change over evolutionary timescales without disrupting the function of that bystander gene.
Similarly, enhancers involved in many biological processes tend to have more-conserved activity. In a study investigating enhancers active in liver tissue across 10 mammalian species, researchers found that, in general, the more traits or cellular contexts in which an enhancer played a role, the more likely it was to have activity in all 10 species.
Scientists continue to investigate what other selective pressures have driven enhancer evolution over hundreds of millions of years and to crack the secrets of the regulatory code. “The field is just in a really interesting place right now, with lots of competing models [and] lots of uncertainty,” Capra says. “And just really cool results.”
Editor’s note: Before work on this article was complete, Jack J. Lee started a communications fellowship at the National Cancer Institute’s Division of Cancer Prevention.








