The past few years have been a stressful time for some of the hermaphroditic nematode worms in geneticist Oded Rechavi’s lab at Tel Aviv University in Israel. Raised at 25 degrees Celsius—a sweltering temperature for C. elegans—the worms have resorted to an unusual coping mechanism: early sex.
Hermaphrodites typically create offspring through self-fertilization until three to four days into adulthood, at which point they start mating with males. But Itai Toker, a former PhD student in Rechavi’s lab, and his colleagues noticed that when the nematodes had been kept in warm conditions for a few generations, they would start to exude male-attracting pheromones just a day after completing their larval development and becoming an adult. That appeared to give them an advantage: When given the choice, male worms tended to mate with these precocious worms more frequently than with ones raised under normal temperatures.
The early sexual attractiveness may be a useful stress response, Rechavi tells The Scientist. More mating means more mixing of different genomes, boosting the progenies’ chances of securing a beneficial allele that will help them weather the taxing environment, he explains. “When you mate, you increase genetic variability” among your offspring.
Continuing to house the nematodes in warm temperatures, the team observed that the premature attractiveness stuck with the worms for at least a dozen generations. Even after the worms were moved back to cooler conditions, the pheromone boost stuck around for three generations, suggesting that the characteristic can be inherited—something Rechavi says he found remarkable. Thanks to their ancestors’ experiences, worms that had never experienced those torrid temperatures still matured earlier. Around the fourth generation in cooler temps, however, worms lost this trait, hinting that there’s a tradeoff to this early mating; sexual outcrossing is generally costly to hermaphrodites as their overall genetic contribution to the next generation is lower, says Toker, who’s now a postdoc at Columbia University. The fact that the early maturation trait was lost also suggested it had an epigenetic basis. Additional experiments showed that small RNAs targeting genes that are involved in sperm development and function were being passed from parent to offspring in the hermaphrodites’ gametes. When the team blocked the transmission of small RNAs, the nematodes didn’t pass on the trait.
The evidence from classical genetics, I should say, is pretty strongly against this being anything at all frequent.
—Brian Charlesworth, University of Edinburgh
Research from other groups has yielded similar evidence that there’s sometimes more to heredity than DNA alone. In C. elegans, the small RNA mechanisms have been shown to transmit acquired traits, including starvation-induced adaptations to food scarcity and avoidance behaviors that prevent the worms from ingesting pathogenic bacteria, even after the initial trigger is gone. “Once it’s going, it seems to just keep going for some time,” says epigeneticist Alyson Ashe of the University of Sydney. Besides RNAs, epigenetic factors such as proteins and other molecules that interact with the genome and direct which genes are expressed have also been found to be passed from parent to offspring in several plant and animal species, sometimes with consequences that affect the fitness of future generations.
Some evidence suggests that epigenetic changes have the potential to influence a lineage’s genetics. Rechavi says that in his study, he’d expect that the nematodes that experienced an earlier start to their mating activity would see an increase in the genetic diversity of their offspring. “This is the punchline of the paper: That a transient small RNA-based response can lead to a permanent change in the genome,” he says. And across a range of species, scientists have learned that extragenomic components can directly change the genetic code by triggering mutations and shaping the evolution of genes. Such revelations suggest that epigenetics could be a long-overlooked player in evolutionary processes, sparking new hypotheses about how species adapt and diversify.
Still, much of the scientific community has expressed doubt that such mechanisms play significant roles in the long-term evolution of populations and species. In many species, eggs and sperm undergo epigenetic “reprogramming,” in which their genomes are largely scrubbed clean of certain epigenetic changes. The uncertainty around the scale and evolutionary importance of epigenetic phenomena is fueling fierce debate about what such findings mean for the foundational evolutionary theories that have shaped science for the better part of the past century. Specifically, the Modern Synthesis developed in the 1940s supposes that evolution is driven solely by random DNA mutations. While many scientists question whether non-DNA-based mechanisms could be meaningful contributors to evolutionary processes, some say that textbooks are due for an update.
“We don’t need to rewrite and throw away the current theories, but they’re incomplete,” says Ashe. “They need adjustment to show how epigenetics can interplay with those theories.”
Interpreting transgenerational epigenetic inheritance
The idea that epigenetic changes can be inherited across multiple generations in animals only began to crystallize relatively recently, with the first case in C. elegans reported in 2006, for instance. Examples of such phenomena have been reported in mice, too, although some studies have suggested that certain types of epigenetic inheritance might be rare in the rodents. Mammalian cells undergo two rounds of epigenetic reprogramming during gamete production and early embryo development, removing the majority of methylation marks from the DNA and reshaping the histone modification landscape, explains molecular biologist Joan Barau of the Institute of Molecular Biology in Mainz, Germany. He adds that in his view, the evidence so far suggests that mammals lack RNA-dependent RNA polymerases (RdRPs), the enzymes that propagate RNA signals across multiple generations in C. elegans. The difficulty in deciphering the mechanism of transmission of epigenetic marks has made it hard to convince many researchers that such intergenerational processes can occur in mammals, says Alexandra Weyrich of the German Centre for Integrative Biodiversity Research Halle-Jena-Leipzig. “The main problem is that we don’t know yet how [epigenetic changes are] transmitted to the next generation. If we solve that, then [the idea of epigenetic inheritance in mammals would be] more accepted [by] evolutionary biologists and geneticists.”
The number of species in which transgenerational epigenetic inheritance has been persuasively demonstrated remains small. Nevertheless, such findings raise an important question, says evolutionary biologist Dragan Stajic of the University of Bern in Switzerland. “If we know now that epigenetic changes can produce variation, and they can be inherited . . . then of course, the logical question is, can evolution act upon this?”
To some scientists, the fact that cases of transgenerational epigenetic inheritance are often ephemeral—typically lasting only a handful of generations in C. elegans, for instance—argues against a significant evolutionary role. For simple traits controlled by single genetic loci, it would be hard for natural selection to produce a population where all individuals bear the same beneficial phenotype if that trait were unstable and reversed in some lineages, says Brian Charlesworth, an evolutionary geneticist at the University of Edinburgh. “That’s one problem, I think, for believing that this is an important evolutionary process—the apparent lack of stability in many of these cases where it does seem to be well-documented,” Charlesworth says.
That’s why some scientists see epigenetic inheritance as no more than an adaptation in and of itself—perhaps a kind of bet-hedging strategy to transiently adapt to short-term environmental stressors without committing to stably transmitted, hard-wired changes. Stajic and colleagues recently tested that idea in the budding yeast Saccharomyces cerevisiae. They engineered yeast strains to have different expression patterns of the uracil synthesis gene URA3 and cultured the fungi in environments that were either changing or constant to see which strains would fare best. The team created the strains by inserting URA3 into areas of the yeast genome known to carry heritable epigenetic factors that cause genes to be either continually expressed or switched on and off at a slow or fast rate. In a regular cell growth medium, the nonswitchers quickly outnumbered the switchers. However, in an experiment that alternated between normal media and one that produces a toxic byproduct when metabolized by the URA3 enzyme—at the same rate as the switchers’ changes in gene expression—the switching strains surged to dominance within 20 generations. “Indeed, we do show that epigenetic inheritance is more beneficial in the fluctuating environments where the conditions change rapidly,” Stajic says. Such mechanisms of adaptation could be of particular importance for species like C. elegans, Ashe adds, which face challenges such as seasonal temperature fluctuations that last only a few months, or a handful of worm lifetimes.
In some ways, whether this kind of transient change amounts to evolution of a population is a semantic issue. Many scientists don’t consider such epigenetically-driven adaptation as evolution, simply because it doesn’t involve changes to coding sequences. “I guess I’m ‘old school’ and would still fall back on the classic definition of evolution as including true genetic change. . . . I’d have to think carefully about the alternate idea of epigenetic ‘evolution,’” says evolutionary geneticist Michael Goodisman of Georgia Tech via email. But to others, such as evolutionary ecologist Christina Richards of the University of South Florida, any kind of heritable change in trait variation should be considered evolution. She points to one of her study species, the invasive knotweed Reynoutria japonica, which has adapted to diverse beach, roadside, and salt marsh habitats. Plants don’t appear to undergo as much epigenetic reprogramming in the germline as animals do, and seem to pass on many methylation patterns quite robustly. If knotweed is acquiring and transmitting epigenetic traits that aid survival in new habitats, she says, “it’s going to affect how well they do; it’s going to affect how much offspring they have. That’s evolution.”
See “DNA Methylation Plays Important Roles in Plant Biology”
Richards adds that with DNA-based changes dominating evolutionary theory for so long, it may well be an uphill battle to convince scientists that other mechanisms contribute. “We’ve been steered by that opinion for a long time,” she says. “If you define evolution as changing allele frequency over time, then we have nowhere to start, and that’s the real problem. . . . I’m not trying to say that it’s all epigenetic. I’m just trying to ask the question [whether it’s involved].”
In fact, inherited epigenetic traits don’t necessarily need to be adaptive to influence evolution, notes behavioral geneticist Benjamin Oldroyd of the University of Sydney. The idea of organisms acquiring an adaptive trait during their lifetime and transmitting it to their offspring—often described as Lamarckian evolution—has long been considered controversial and such phenomena have only been observed in organisms like C. elegans and potentially in certain plants. All it takes for epigenetics to affect evolution, Oldroyd says, is a change in gene expression—triggered, for instance, by an environmental perturbation—and for those epigenetic marks to be inherited. Moreover, Stajic adds, if some organisms habitually pass down certain traits epigenetically, as do many plant species, then any random errors that occur during the transmission of those marks could produce new heritable epigenetic traits. In theory, natural selection could act on all these types of epigenetic changes, Stajic says.
Some researchers note that the evidence for transgenerational epigenetic inheritance—and especially its role in evolutionary processes—outside of laboratory organisms remains thin. In plants, for instance, which have been eagerly studied for such effects, “The most we can say is that we have clues that epigenetic mechanisms might have a role in adaptation,” says plant ecologist Teresa Boquete of the Universidade de Santiago de Compostela in Spain. Many studies in wild plant species have been limited to describing broad epigenetic differences across individuals, and it’s often unclear whether these changes have taken place within an organism’s lifetime or if they’ve been inherited across generations, she says. Another challenge with wild populations that aren’t genetically identical is that the observed patterns may not be purely epigenetic, if there are underlying mutations in the genes encoding the organism’s epigenetic machinery, Richards notes. “Even one change in DNA sequence can dramatically change the methylation patterns in the genome.”
In humans, the evidence is even harder to interpret, with correlational studies so far failing to yield definitive answers about transgenerational epigenetic inheritance, Barau says. “I’m not a total [skeptic] of transgenerational epigenetic inheritance, it’s just that the burden of proof for this is quite high. And it hasn’t been achieved in many of the [claims], especially when you’re talking [about] humans.”
See “Does Human Epigenetic Inheritance Deserve a Closer Look?”
Epigenetics and the GenomeScientists are learning that epigenetic marks such as DNA methylation can influence not just gene expression, but genes themselves. When this happens in germline cells, the changes can diversify the genetic substrate upon which natural selection acts. Scientists are still exploring how such processes could influence evolutionary processes overall. |
Triggering mutations Scientists have known for several decades that methylation can cause the nucleobase cytosine to lose an amino group, transforming it into thymine. When this occurs during gametogenesis, it can be one of the major drivers of evolution at the molecular scale. Some human embryo studies estimate that for particular genomic regions, some 20 percent of mutations unique to humans arose from cytosine methylation. |
Shaping mutation patterns Some studies in cancer cells suggest that the packaging of DNA into dense structures called nucleosomes not only shapes the expression of those genes, but also their mutation rates. When bound in such conformations, DNA is less likely to open into a single-stranded state that makes it more prone to mutations. |
Dampening mutation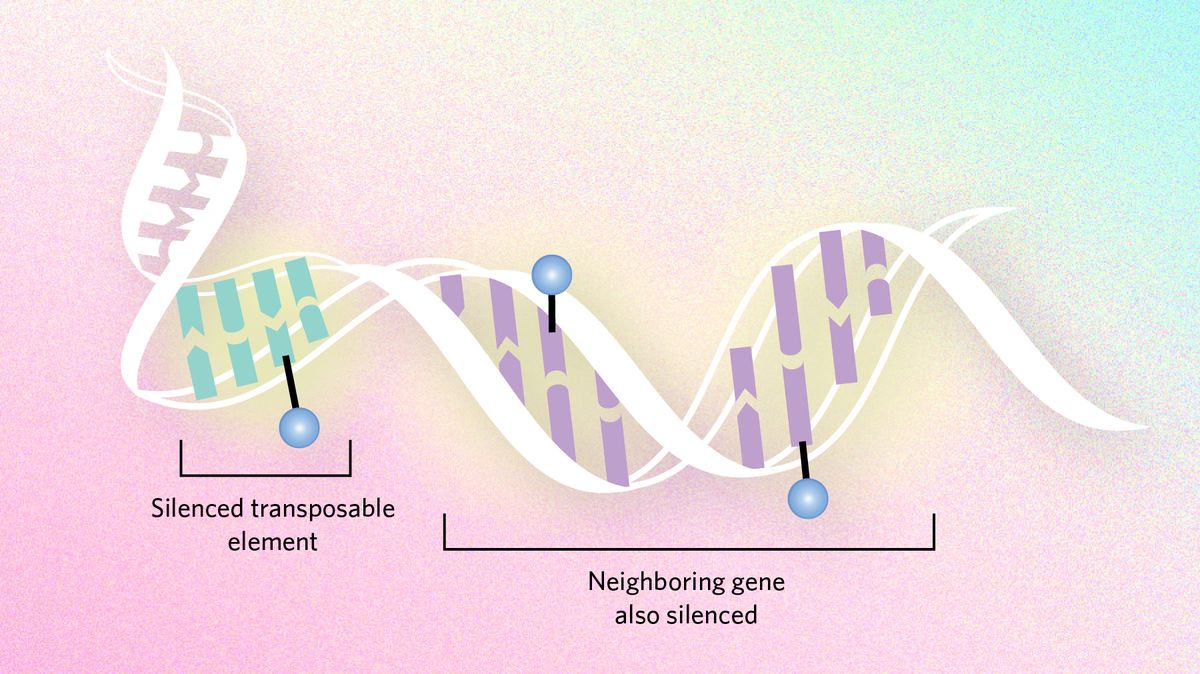 Epigenetic dynamics can also slow evolutionary processes by suppressing transposable elements, snippets of DNA that hop around the genome and cause mutations. Genes neighboring these troublemakers can end up being silenced too, sometimes creating a conflict between the need to express genes and the need to silence transposons. In certain Drosophila species, researchers hypothesize that the flies have moved large chunks of the transposon-infested Y chromosome to other chromosomes to ensure important genes are not silenced. |
New gene functions Long-term epigenetic silencing of particular genes could allow them to accumulate genetic mutations and develop new functions over time. There is some correlational evidence that duplicated genes—which are often lost from the genome unless they are beneficial to the organism—frequently become epigenetically silenced. They could thereby evolve new functions. |
Effects of epigenetics on the genome
Given the often short-lived nature of epigenetic inheritance, one way that inherited extra-genomic factors could influence evolution is by changing the DNA itself—a phenomenon for which there is some evidence now surfacing. (See illustration.) Rechavi’s group has shown this can occur for an animal, albeit indirectly, by promoting sexual reproduction in the worms and thereby turbocharging their genetic evolution. In that case, the mutations that make their way into young worms’ DNA through genetic recombination are random and scattered across the genome; the effect of the epigenetically inherited trait is simply that their general genetic diversity is increased—although Rechavi adds that the team didn’t sequence the offspring’s genomes to explore how exactly this ultimately affects genetic evolution.
Another hypothesis that some scientists have floated in the literature is that inherited epigenetic regulation of particular genes can influence genetic adaptation at those specific genes. If an environmental pressure maintains long-term gene silencing, for example, the gene could naturally accumulate random mutations over time, changing its function or, more likely, rendering it defunct. At some point, “that epigenetic silencing of that gene would no longer be required because the gene has mutated anyway,” Ashe says.
That idea hasn’t yet been tested in animals, Ashe cautions. But a 2019 study by Stajic and colleagues provides some proof of concept in yeast cells that had been engineered to have URA3 partly silenced. When the researchers placed the strains in a potentially toxic growth medium, “We actually see that this first step in adaptation is through epigenetics,” Stajic says. Specifically, the cells strengthened silencing of URA3, and some generations later, knockout mutations popped up that rendered the gene defunct. In this way, epigenetic inheritance could form a kind of bridging mechanism to long-term change, Stajic says—in essence, a “soft” adaptation to probe the waters, followed by a hard-wired change that will stick in the genome indefinitely. “It seems that epigenetics, in this case, acts as a buffering system that allows the population to survive this initial step . . . until the beneficial mutation appears and fixes,” he says.
We don’t need to rewrite and throw away the current theories, but they’re incomplete.
—Alyson Ashe, University of Sydney
More directly still, some epigenetic marks have long been known to induce mutations and shape mutational patterns across the genome. Methylation marks, for instance, are known to trigger a chemical reaction that mutates cytosine into a different base, thymine. Methylation patterns also influence genomic stability by repressing highly mutagenic DNA snippets called transposable elements. “These stability changes can influence the DNA sequence directly, which subsequently can lead to genetic adaptation,” Weyrich says. A recent analysis of the Arabidopsis thaliana genome suggests that mutations aren’t truly random but are shaped by epigenetic marks, including not just methyl groups but also histone modifications and the general accessibility of chromatin. When such mutations occur in germline cells and become inherited across generations, they have the potential to influence a species’ evolution, Goodisman says.
Through mechanisms like these, says evolutionary biologist and philosopher of biology Eva Jablonka of Tel Aviv University, epigenetic inheritance could even lead to the diversification of species. “The first thing that happens in an environment is an adaptation at the physiological level,” she says, “and then anything that helps [the organism] to get stabilized will be selected.” So far, there is only correlative support for this idea. She points to research in darter fish (genus Etheostoma) suggesting that methylome differences are greater than genetic differences between two closely related but physically separated fish populations; the authors hypothesize that epigenetic differences could be driving reproductive isolation between them. Similar observations have been made in populations of the spiny mouse Acomys cahirinus and in farmed breeds of the European sea bass Dicentrarchus labrax. In the sea bass, researchers even found that epigenetic differences identified in early domesticated populations overlapped with cytosine-to-thymine mutations documented in samples that had undergone 25 years of selective breeding. And, the fact that phenotypic diversity between species is greater than can seemingly be explained by genome differences alone—with just a few percent sequence difference between chimpanzees and humans, for instance—hints that factors other than DNA play a role in species diversification, Weyrich adds. “If we exclude this flexible mechanism completely . . . then you just miss a very important piece of evolution and of diversity.”
The role of epigenetics in speciation is still debated. “I’m not saying there isn’t something there, but . . . solid evidence is really lacking,” says Charlesworth. That’s perhaps because such evidence is simply hard to get, he notes. It’s difficult to robustly quantify epigenetic changes between populations, because epigenetic patterns vary so much between individuals and even within an individual’s lifetime. Compared to genetic mutations, which can be quantified with reasonable accuracy, “when it’s something like the amount of methylation, it’s a much vaguer sort of statistic.”
A future full of questions
While many questions surrounding epigenetic inheritance remain unsettled, Jablonka sees the evidence accumulated thus far as a validation of hypotheses she and others conceived of decades ago. She and evolutionary biologist Marion Lamb saw the potential of epigenetics to influence evolutionary processes in the 1980s after separately studying epigenetic processes in cells of different rodent species and fruit flies. The pair conceptualized epigenetic inheritance as another evolutionary driver, alongside genetic change and the behavioral and cultural transmission of traits. All four pathways can generate heritable variation on which natural selection acts, they wrote in their 2005 book, Evolution in Four Dimensions.
These notions are key to the Extended Evolutionary Synthesis (EES), a theoretical framework that aspires to be a complement to the DNA-centered views laid out in the Modern Synthesis. In the EES view, evolution occurs not solely by changes in genetic allele frequency, but via any kind of heritable variation in traits—a view that Jablonka sees as increasingly supported. “The more people look, the more they find.”
Charlesworth, however, doesn’t see epigenetic inheritance as a challenge to fundamental rules of evolution laid out in the Modern Synthesis. Many documented cases involve subtle traits pertaining to stress responses and fertility, and not lineage-defining evolutionary innovations such as eyes or wings, for instance. In his view, it’s likely that the documented cases of epigenetic inheritance in worms, plants, and mammals are exceptions to the normal rules of heredity, he says. “The evidence from classical genetics, I should say, is pretty strongly against this being anything at all frequent. People have been working on genetics for over 120 years. There’s a huge body of evidence on . . . how [traits are] inherited.”
Such discussions tap into a broader argument that has long raged over the EES, with frustration on both sides, researchers tell The Scientist. A recent article in The Guardian titled “Do we need a new theory of evolution?” generated rigorous debate among scientists on Twitter. “This all brings very heated discussions [at] the conferences and meetings,” Stajic says via email. “It is definitely very exciting times to be in the field.”
The outcome of this rift—as least as far as the evolutionary role of epigenetics is concerned—hinges in part on the continued hunt for such phenomena. To that end, Stajic is planning a study on stress adaptation in zebrafish, while Richards says she’ll continue to probe knotweed for epigenetic effects. Rechavi, meanwhile, hopes someday to scrutinize wild isolates of C. elegans or of other worm species to see if they behave anything like their laboratory counterparts. He finds it hard to believe that the observations made so far are just a coincidence. “My feeling is that it’s quite unlikely that we happen to find it in just [a few] of the . . . model organisms of biology and it doesn’t exist in any other organism,” he says. “This seems like too much luck.”

