Light microscopes are quintessential icons of the biological sciences, critical infrastructure in nearly all life science laboratories. Every cell culture lab has one, as do high school teaching labs; for less than $100, you can pick one up at your local science or toy store. For many applications light-microscope technologies are more or less routine, and the systems that enable them are essentially turnkey—which might suggest that light microscopy has plateaued, technologically speaking. Nothing could be further from the truth.
Researchers on the cutting edge are constantly pushing the limits of the instruments’ optics, seeking to extract ever-sharper images from increasingly demanding samples, all while inflicting the least possible photodamage. From live-cell imaging to confocal microscopy, super-resolution imaging to multiphoton excitation, new microscopy technologies and applications appear regularly in the literature.
The Scientist asked four microscopy innovators to talk about the novel applications and approaches they developed in their...
CHASING (FRUIT) FLIES
Researcher
Philipp J. Keller, Fellow, Howard Hughes Medical Institute, Janelia Farm Research Campus, Ashburn, VA
Project
Mapping cell migration during Drosophila embryogenesis
Problem
In 2008 Keller published a technical tour de force in which he tracked cell migration over 24 hours of zebrafish development (Science, 322:1065-69). But that approach won’t work for fruit flies, he says—they are too opaque to image easily.
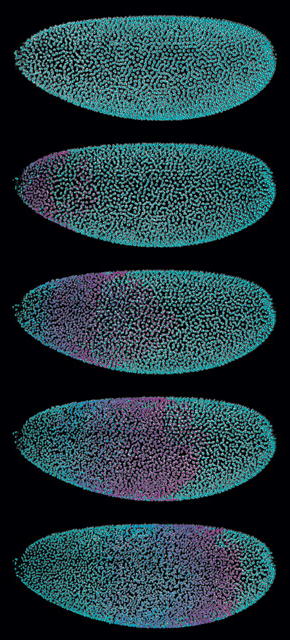
Solution
Keller’s 2008 study used light-sheet microscopy, a technique that rapidly scans a sample from the side with a thin plane of laser light and images the resulting fluorescence at a right angle. The approach enables researchers to make relatively long-term recordings with minimal photobleaching and phototoxicity.
But the approach does not work well with larger samples (such as mouse embryos), says Keller, nor is it compatible with relatively opaque samples, such as Drosophila. To circumvent those issues, Keller’s team developed a new system called SiMView (simultaneous multiview light-sheet microscopy), which uses four objectives—two for excitation and two for detection—to record four complete and simultaneous data sets, in either single-photon or multiphoton modes. “Each combination of illumination objective and detection objective . . . provides a different and complementary view of the sample,” Keller explains.
According to Keller, the system can collect a data set of 150 optical slices through a Drosophila embryo in each of four directions—600 images total—every 5 to 10 seconds. That’s about 6 gigabytes of data per 3-D “volume,” which the system then processes and collects repeatedly over the course of the experiment, tracking cellular activity in real time. In one image series, the system captured a front of mitotic nuclei as it rippled across the embryo from anterior to posterior over 3 minutes, a magenta wave of cell division on a cyan background.
Naturally, the images pile up fast. A complete 20-hour data set amounts to a whopping 10 to 15 terabytes of data. That’s about as much as all the text housed in the print collections of the Library of Congress, Keller says. “That would be the equivalent to one Drosophila time-lapse recording.”
DIY
According to Keller, light sheet–based systems are relatively inexpensive to build. The key, he says, is a good detector (or for SiMView, two), laser unit, and optics. “The rest is basically just a couple of mechanical parts.” He estimates that his 2008 system could be built for between $30,000 and $50,000, while a SiMView scope might cost around $150,000. That’s just half the price of many commercial confocal systems, he notes, which can cost upwards of $300,000.
CORRECTING ABERRATIONS
Researcher
Martin Booth, Advanced Research Fellow, Centre for Neural Circuits and Behaviour, University of Oxford, U.K.
Project
Imaging Drosophila brains during learning and behavior
Problem
Biological specimens are not uniform. Differences in optical density in different parts of the specimen can distort the resulting images, especially in thick specimens.
Solution
The problem of image distortion is familiar to astronomers; it occurs as starlight passes through the Earth’s atmosphere. Astronomers correct for that using adaptive optics—a sensor that detects the distortion and a deformable mirror that reverses it, producing a sharp image.
Booth uses what he calls a “sensor-less” adaptive optics strategy that instead bends the microscope’s mirror into specific shapes to find the one that best corrects for the distortion in his images. He implemented the strategy in a custom-built, two-photon microscope, which further improves image clarity by limiting fluorescence occurring outside the focal plane. Booth’s goal is to apply this technology to imaging neuronal signaling in vivo. Earlier this year, he took a step in that direction when he coauthored a study describing a new imaging system that increases the speed of fluorescent scanning of fixed mouse cortical slices in all three imaging directions to about 3,000 scans per second, about 300 times as fast as the original system (PNAS, 109:2919–24, 2012). Traditionally, scanning in the plane perpendicular to the imaging objective is fast, but axial scanning is relatively slow. “Neurons rarely lie in the perpendicular plane, so you need to be able to scan in all directions at high speed,” he explains. This new system enables that by employing a lens and a fast-moving mirror, which rapidly tracks fluorescence along neuronal processes, such as axons.
Now Booth is applying that approach to live fruit-fly brains as the flies learn and respond to stimuli. Using this new system, he hopes to achieve the same high resolution that confocal and two-photon systems can provide of fixed samples on microscope slides—but do it in live animals over time. “Ultimately, that’s where we want to be doing biology,” he says.
DIY
There are no commercial adaptive optics microscope systems currently available, Booth says, but you can build your own. Expect to pay $20,000 to $40,000 for the parts, plus whatever your time is worth to assemble the system, write your own controller software, and work out all the bugs.
DEEP-TISSUE SUPER-RESOLUTION
Researcher
Hari Shroff, Chief, Section on High Resolution Optical Imaging, National Institute of Biomedical Imaging and Bioengineering, Bethesda, MD
Project
High-resolution imaging of zebrafish embryonic development
Problem
Super-high-resolution microscopy techniques typically cannot be applied to live samples, because they are both slow and relatively toxic to the cells. The exception is SIM (structured illumination imaging), which has now been commercialized by Nikon and Zeiss. But even SIM is reserved mostly for single-cell experiments.
Solution
Shroff built a system that couples the super-resolution of SIM—a technique that basically halves the diffraction limit of light—with the ability of confocal microscopy to reject out-of-focus light in thick samples, allowing him to image samples several cell layers deep. The resulting instrument, called a multifocal structured illumination microscope (MSIM), uses a digital micromirror device (DMD)—the same component that drives digital movie projectors—instead of the spinning-disk pinhole system found in many confocals. “We had a DMD lying around, and we didn’t have a spinning-disk confocal scan head,” he says, adding that the approach would probably also work with a spinning-disk confocal unit “with the right modifications.”
Shroff’s team can now image samples at least 8 times thicker than traditional SIM systems, up to 50 or 60 microns thick. In one analysis, the team took a live zebrafish embryo and collected serial optical slices starting 10 microns beneath the surface. In total, they collected 22 optical slices, each 0.6 microns thick, repeatedly for about 50 minutes. The resulting data illuminate the mechanisms underlying development of one of the fish’s sensory organs.
DIY
MSIM is simpler to implement than other SIM configurations, Shroff writes in the paper describing this technology (Nat Meth, 9:749-54, 2012). “We add only a DMD, a telescope, and a fast camera to a conventional wide-field microscope. Our illumination path has no moving parts and is insensitive to polarization.”
MINDING THE PHOTON BUDGET
Researcher
Eric Betzig, Group Leader, HHMI Janelia Farm Research Campus, Ashburn, VA
Project
Imaging dynamic processes on a cellular scale
Problem
As the spatial and temporal resolution of microscopy studies increases, the number of fluorescence photons available for detection—what Betzig calls the “photon budget”—decreases. “An increasingly important goal,” he says, “is how to use that photon budget as wisely as possible.”
Solution
Like Keller, Betzig uses light sheet–based microscopy for its speed, long-term imaging, and low phototoxicity. But Keller’s light sheets, which work well enough for tracking nuclei in intact embryos, are too thick to resolve subcellular details.
Generally, Betzig says, there’s a relationship between a light sheet’s flatness and how thick it is: to make the sheet as flat as possible, it must also be relatively thick—a big, flat textbook say, instead of a thin but rippled pamphlet. Betzig circumvented this problem using an optical trick called a “Bessel beam” to produce a far thinner “pencil” of light, which he then sweeps back and forth across a single-cell-size sample to create a plane.
“What we’re doing is basically a wrinkle on plane illumination to make it suitable for doing high-resolution imaging of single cells,” he says.
The resulting sheets can be as thin as 0.3 microns, as opposed to traditional sheets, which measure about 3–8 microns thick. But Bessel beams have a wrinkle of their own, Betzig notes: the pencil’s cross-section actually resembles a bull’s eye, with a sharp center and progressively dimmer concentric rings. Those rings can induce fluorescence, too, reducing image clarity. So the team developed two solutions, one using two-photon imaging to eliminate out-of-focus light and the other involving a super-resolution system that uses the rings to enable a form of SIM.
Employing these different approaches, Betzig’s team recorded filopodia motion on HeLa cell surfaces, tracked membrane dynamics on monkey kidney cells, and monitored chromosome movement during mitosis—for up to 40 minutes or more in some cases. In the case of chromosomal tracking, the team captured a stack of 200 two-dimensional planes per second (with variable rest intervals) over at least 10 minutes, collecting a total of 200 volumes. That’s some 40,000 image frames, he notes, with no apparent cellular damage (as mitosis continued unimpeded).
DIY
None of the major microscopy vendors offers a commercial light sheet–based microscope, let alone one with Bessel imaging. But you can make one yourself. “If you’re comfortable with building your own confocal microscope, you should not have any trouble building a Bessel microscope,” Betzig says. He estimates the cost at around $150,000 to $200,000. His lab even offers schematics and computer code, provided researchers sign a nondisclosure agreement.
Interested in reading more?




